Blog
Home
The Past, Present, and the Future of DNA Computing
I would be giving my Prelim (which essentially graduates me into a PhD candidate from a PhD student) in a couple of weeks and I am having serious questions about my intellect and my capability of finishing a PhD (and to be honest, surviving the prelim). But I somehow have to prepare some slides and pen a 30-page document, whilst I try to address the comments of a harsh reviewer who hated my first paper :3. Anyhow, I would be reading a lot of papers while preparing my slides. So I thought, why not compile them for the future, more intelligent MEs to peer at, and be less anxious! Maybe, this will motivate me to read the paper till the end. So, here goes.
Seesaw gates
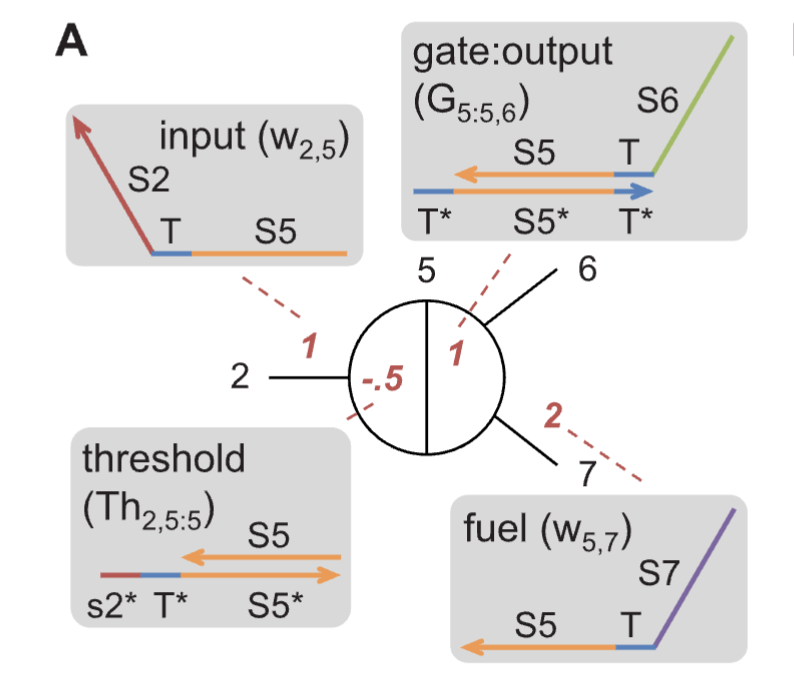
[1] introduced the design of seesaw gates in the context of boolean logic computation. Each of these gates contain a base strand containing a “recognition” domain flanked on either sides by two toeholds. One of these toeholds is free, whereas the other is occupied by an incumbent signal strand. Seesaw gates enable catalytic signal transduction. The addition of the input displaces this outgoing strand, occupies the recognition domain, and opens up the previously bound toehold. A fuel strand then occupies this free toehold displacing the input back into its single-stranded form. Here, it is worth noting that each signal strand is designed such that it has a toehold in the middle, surrounding on the either side by recognition domains. Each of these recognition domains represent the gate they are returning from and the gate they are impinging upon, respectively.
In the first stage, catalytic amplification and thresholding happens. A point to note is that these gates assume that the thresholding happens much faster than the strand displacement (increases the toehold length of $Th_{2,5:5}$ complex). The gate operation happens in two stages using two different seesaw gates. In the first stage, the output is generated and thresholded. In the second stage, the generated outputs from each of the upstream gates is integrated at the “Integration” gates. The final output is again thresholded and the rest is sent to the reporter for output generation.
By imposing the concentrations of the gates to be $1$ and the threshold to be around $0.6$, OR gate could be simulated and with a threshold of $1.2$, AND gate can be simulated. Note that at each gate, an output stoichiometrically equivalent to the gate complexes is produced.
Possible improvements
- Can the gates be replenished somehow? Currently, they are one-shot devices.
- Can we somehow forego the fuel complexes and figure out a way to implement automatic buffering?
- How can we speed up computation?
- Also check: [1], [2], [3], [4], [5]
Reaction-Diffusion patterns in DSD-based CRNs
Dalchau, Seelig, and Phillips (2014) designed an extension for the VisualDSD framework to include spatial patterning by implementing PDE evaluators. They modeled the spatial patterns formed by various dynamical systems including, autocatalytic amplifier, Lotka-Volterra oscillator, and a consensus system. They simulated these systems both in the presence and absence of leaks.
In the autocatalytic amplifier, they observed travelling waves in the absence of leaks. However, they observed that this patter is highly sensitive to leak.
In the Lotka-Volterra oscillator, they observed periodic oscillatory waves, albeit with decreasing amplitude. Figure below shows these patterns.
More stable, equilibrium patterns were observed in the reaction-diffusion system of a majority consensus system. Remarkably, these patterns were qualitatively unaffected by leaks, as is the case with consensus algorithms.